RNA interference (RNAi)
As well as producing messenger RNA, DNA in fact also encodes other forms of RNA that are involved in cell regulation.
These are transcribed by
the same form of RNA polymerase as the enzyme that is involved in building up mRNA - or more accurately pre mRNA. Other forms of RNA polymerase are involved in the transcription of
tRNA and
rRNA .
This underlines the notion that DNA codes for more RNA than the mRNA that is translated into protein.
Small nuclear RNAs (snRNAs) operate within nuclei, where they are tightly bound to proteins to form small nuclear ribonucleoproteins, or snRNPs (often pronounced snurps), that control the splicing of pre-mRNA.
See Action of spliceosomes opposite
Translation of the mRNA produced from target genes can be inhibited by RNA interference (RNAi). This is called
silencing the gene.
MicroRNA 'stem loops'

The first miRNA discovered (in 1993), lin-4 from
Caenorhabditis elegans is an endogenous regulator of genes that control developmental timing.
MicroRNAs (miRNAs) are formed as hair-pin bends of single-stranded RNA that fold back on themselves to give 'stem loops' about 22 to 26 nucleotides long.
One section of this becomes incorporated into a protein-based
RISC (RNA-induced silencing complex).
Small interfering RNAs (siRNAs) are formed as double-stranded molecules about 21 to 25 base pairs long. One of the strands becomes incorporated into a protein-based RISC.
When the single-stranded miRNA or siRNA within a RISC binds to a molecule of mRNA that contains a sequence of bases complementary to its own, the mRNA is either hydrolysed or its translation is stopped.
This can be used to degrade mRNA that is no longer needed to produce protein.
It can also act as a defence against infection by viruses.
Action of spliceosomes
Pre mRNA, produced by transcription in the nucleus, consists of exon sections alternating with intron sections.
Each of the intervening intron sections are removed by the action of 'spliceosomes', composed of small nuclear ribonucleoproteins - complexes containing catalytic proteins assisted by sections of RNA. Only the exons will be expressed after the splicing process
Structure of U1 snRNA

There are several varieties of small nuclear ribonucleic acid (
named U1 - U6) and they consist of double-stranded sections, held together by complementary base pairing, with loops, exposing single stranded sections, like transfer RNA. This enables them to target the two ends of each intron, and presumably to bring in the appropriate proteins.
Different snRNAs become attached to the GU nucleotide sequence at the 5' end splice site, and the AG sequence at the 3' end splice site, and these are then brought together as the intervening section is formed into a loop - often described as a 'lariat' - which is cut out as two exon sections are bonded together.
Mature mRNA will eventually be expressed by being translated into protein at the ribosomes in the cytoplasm, and the intron lariats are broken down into RNA nucleotides.
Post-transcriptional gene silencing
This enables removal of mRNA - either originating in the cell (and no longer needed?) or RNA from pathogens such as invading viruses.
Alternatively RNA may be introduced into the cell in an attempt to treat genetic conditions.
siRNA and miRNA are processed so as to form a template to bind 'target RNA' to be broken down by various protein complexes.
Action of Dicer
Silencing of mRNA by RNA interference

This enzyme breaks down double-stranded RNA (dsRNA) and pre-microRNA (pre-miRNA) into short double-stranded RNA fragments about 20-25 base pairs long, with
two exposed single stranded bases at the 3' ends -
small interfering RNA and
microRNA.
These fragments are surrounded by a number of proteins to form the RISC. One strand - the 'anti-sense' or 'guide' strand - is retained, and the other 'sense' or 'passenger' strand is degraded.
RNA-induced silencing complex, or RISC
This consists of several proteins, arranged around the short section of RNA, a projecting part of which acts as a template that recognises mRNA to be disposed of. This is attracted as a result of complementary base pairing between the RNA bases.
Several of the proteins are
RNA-specific endoribonucleases
The protein called
Argonaute (Ago) breaks down the mRNA (attached to the anti-sense strand) into short sections of nucleotides.


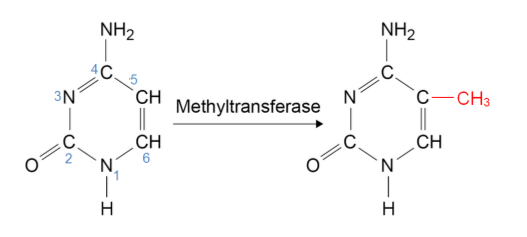

 As N replaces C at position 5, this (base) does not have a site for methylation.
As N replaces C at position 5, this (base) does not have a site for methylation.
 The first miRNA discovered (in 1993), lin-4 from Caenorhabditis elegans is an endogenous regulator of genes that control developmental timing.
The first miRNA discovered (in 1993), lin-4 from Caenorhabditis elegans is an endogenous regulator of genes that control developmental timing.

