The Sanger or chain termination procedure
There are some points of amplification, and questions to test your comprehension in the section on the opposite side, (or beneath)
This technique requires four reactions to be carried out at the same time, in separate tubes.
All 4 tubes contain
- a large quantity of the sample DNA, in the single strand form
- a large quantity of the four nucleotides containing thymine, cytosine, guanine and adenine
- DNA polymerase enzyme
- radioactive primers
- To each tube is also added a lesser amount of a different (single)
modified nucleotide - which lacks a -OH group on the deoxyribose section. This may also be called a '
terminator base', although it is obviously bonded to (modified) deoxyribose and phosphate(s).
The primers act as starter points for the attachment of DNA polymerase, which adds nucleotides to the second strand (rebuilding the double helix).
However once a modified nucleotide is added, the DNA polymerase cannot make a phosphodiester bond in the outside edge of the developing DNA strand, so the chain is terminated.
It has been suggested that the modified nucleotide does not "fit" the active site of DNA polymerase, but the condensation reaction forming the phosphodiester bond needs an -OH group.
The two DNA strands are separated again. There will be a range of strand lengths. Half will be complete (original strands), and half will be of variable length - probably incomplete due to chain termination. On the end of these will be the modified nucleotide added to that tube.
The contents of each tube are then loaded into wells in a film of polyacrylamide urea gel which does not allow the DNA strands to anneal.
This gel layer is then subjected to electrophoresis to separate the components according to their molecular size. It is then overlaid with photographic film and this is 'developed' after some time to give an autoradiograph.
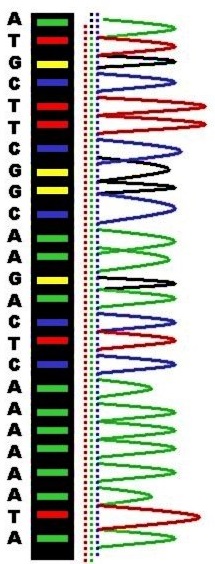
Autoradiograph
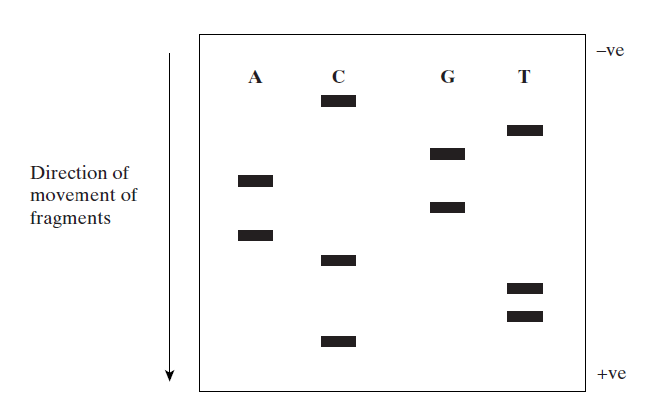 Letters indicate the modified nucleotide in each tube
Letters indicate the modified nucleotide in each tube
The sequence of bases in the DNA can be read from the autoradiograph. Smallest sections of DNA travel furthest, so the lowest bars show the first bases in the sequence.
This sequence is CTTCAGAGTC
so the original strand was
>
GAAGTCTCAG
What causes the DNA fragments to move in the electrophoresis process?
>
Negatively charged phosphate groups (on the outside of the helix) are pulled towards the anode (+ve electrode)
Single nucleotide substitution revealed by sequencing
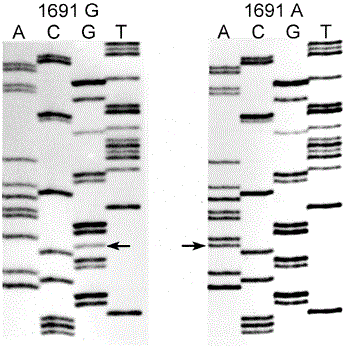 (Section of)
(Section of)
Normal allele on the left, mutant allele on the right
From Wikimedia Commons, the free media repository
I noticed this pair of autoradiographs when searching for another topic.
It ties up with the topic of Base sequence alteration on this site (you can check the genetic codes there, too).
Factor V Leiden is the most common hereditary blood clotting disorder amongst ethnic Europeans.
Factor V is a cofactor allowing factor Xa to activate prothrombin, resulting in the enzyme thrombin which in turn cleaves fibrinogen to form fibrin, forming fibres that result in a clot. This clotting process is then controlled by (activated) protein C - a natural anticoagulant that acts to limit the extent of clotting by cleaving and degrading Factor V.
The mutant ('Leiden') form of the gene reponsible for this protein is caused by a 1691G → A substitution (shown by arrows). In fact reading up from one below the arrow to one above it you can see the triplet of bases is
CGA on the left, whereas on the right the triplet is
CAA.
This results in a change in the amino acid sequence of the protein coded for by the gene - changing a single amino acid arginine R to a glutamine Q. This change in structure prevents the cleaving action of protein C so that any events causing blood clotting can have more serious consequences than usual.
More details, and a few questions about the chain termination procedure
The sample DNA is amplified using the PCR process, and 'denatured' - so it is in a single-stranded form.
How is the sample DNA converted into a single-stranded form?
>
heated - temperatures over 90 °C prevent hydrogen bonds forming
The nucleotides are actually triphosphates dATP, dGTP, dCTP, and dTTP.
What is the significance of the nucleotides being in the form of triphosphates?
>
This gives energy (think of ATP) for the formation of phosphodiester bonds which hold the backbone together
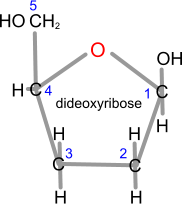 Dideoxyribose has no -OH groups at positions 2 and 3
Dideoxyribose has no -OH groups at positions 2 and 3
The modified nucleotides are in fact
dideoxynucleotides (ddNTPs - ddATP, ddGTP, ddCTP, or ddTTP). They do not have an -OH group (just 2 -H) in position 3 of the deoxyribose, so it is not possible to form a phosphodiester bond, and the DNA backbone of the developing second DNA strand will stop here.
Why is only a small quantity of dideoxynucleotide needed?
>
It is only needed once in each tube, whereas the other ordinary nucleotides are needed to fill in at positions leading up to that
Primers migrate into position alongside the 3' end of the single DNA strands, and bind ('anneal') to it.
What causes this binding process?
>
hydrogen bonding, using complementary base pairing - NOT phosphodiester bond
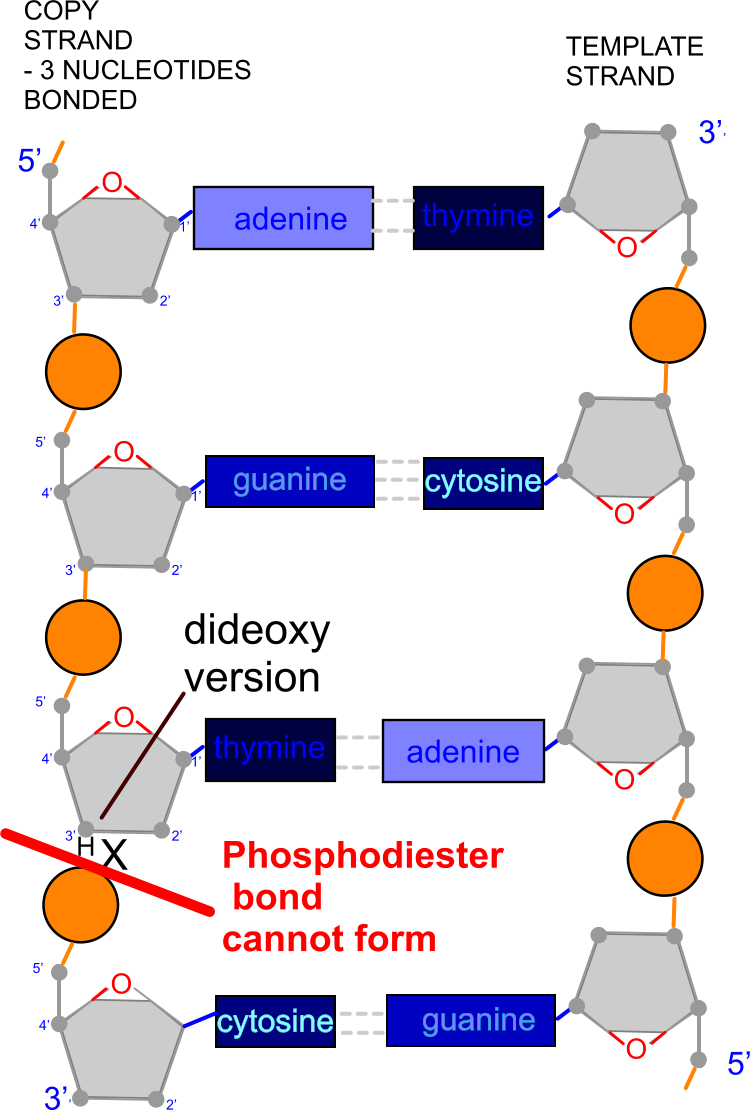
However once a dideoxynucleotide has paired up with its complementary base on the DNA template, the DNA polymerase cannot make a phosphodiester bond between the carbon at position 3' and the carbon at position 5' on the next nucleotide, so the (second)
chain terminates at this point.
In the diagram alongside on the left three nucleotides have successfully formed a chain alongside the template strand, but the next nucleotide (with the base cytosine) cannot join on because the nucleotide above it (with the base thymine) has no -OH group at 3' to form the phosphodiester bond (just -H).
Polyacrylamide urea gel does not allow the DNA strands to anneal, and prevents single-stranded DNA forming into a circular form.
Suggest how this binding process is prevented.
>
Urea prevents hydrogen bonds forming between bases.
What causes the bars seen on the autoradiograph?
>
radioactivity (in the primers)
Elements used in radioisotope labelling include sulphur (Sulphur-35), phosphorus (Phosphorus-32) and iodine (Iodine-125).
Which of these would be used in this case?
>
phosphorus - which is in the 'backbone' of the DNA
Alternatively, DNA fragments may be visualised using a variety of dyes.
Ethidium bromide migrates into the gaps between bases and it can be seen when illuminated by ultraviolet light. It is considered to be a potential carcinogen so it is not used in educational contexts. Other coloured chemicals may be used, however.
 from the Nobel Prize website
from the Nobel Prize website
 Letters indicate the modified nucleotide in each tube
Letters indicate the modified nucleotide in each tube

 Dideoxyribose has no -OH groups at positions 2 and 3
Dideoxyribose has no -OH groups at positions 2 and 3
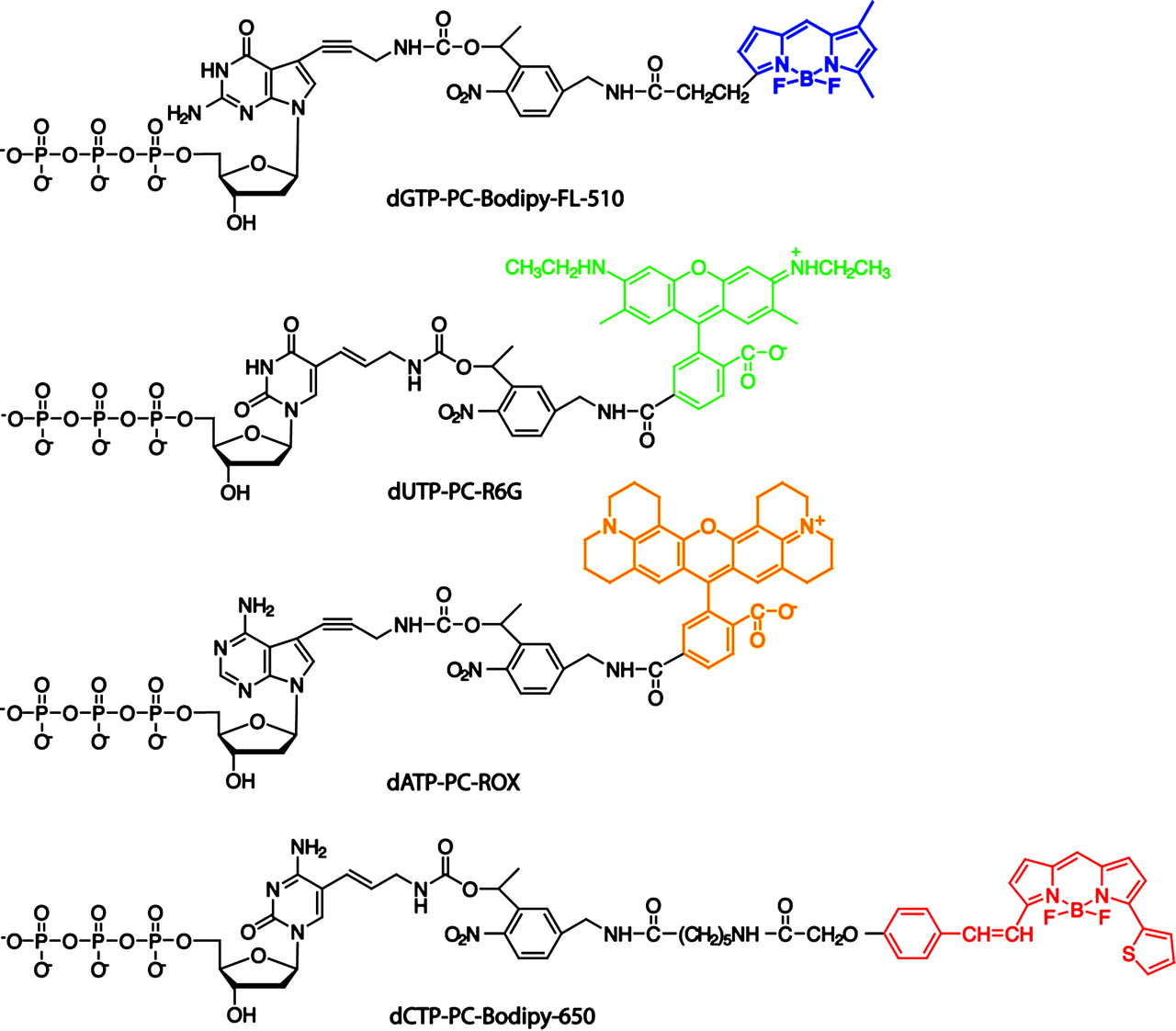 The ring-shaped sections fluoresce with different colours when illuminated by laser light.
The ring-shaped sections fluoresce with different colours when illuminated by laser light.